23 -- Cascaded Detector Performance Analysis with an Application to Cell Detection
Thomas Athey, Nir Shavit
As both computer vision models and biomedical datasets grow in size, there is an increasing need for efficient inference algorithms. We utilize cascaded detectors to efficiently perform object detection of sparse objects in multiresolution images. Given an object's incidence and a set of detectors at different resolutions with known sensitivities and specificities, we derive the sensitivity and specificity of the associated n-level cascaded detector. We also derive the expected number of detector executions in a two-level cascade detector and analyze its dependence on various parameters. Finally, we compare one- and two-level random forest-based detectors in a cell detection task in three-dimensional sparsely labeled fluorescent neuron images. We show that the multi-level detector shows comparable performance in less than half the execution time on multiple datasets. We believe that our work can be extended to detect sparse objects in a variety of biomedical data domains and signal dimensions.
24 -- Rapid Epigenomic Classification of Acute Leukemia
Salvatore Benfatto, Til L. Steinicke, Maria de los Reyes Capilla Guerra, Andre B. Monteleone, Evan C. Chen, Gabriel K. Griffin, Volker Hovestadt
Acute leukemia (AL) is an aggressive form of blood cancer that requires precise molecular classification and urgent treatment. However, standard-of-care diagnostic tests are time and resource intensive and do not capture the full spectrum of AL heterogeneity. Here, we developed a machine learning framework to rapidly classify AL using nanopore-based genome-wide DNA methylation profiling. We first assembled a comprehensive reference cohort (n=2,540 samples) and defined 38 distinct methylation classes across AL lineages and age groups. Methylation-based classification closely matched lineage classification by standard pathology evaluation in most patients and revealed disease heterogeneity beyond that captured by standard genetic categories. Using this reference, we developed a specialized deep neural network model (MARLIN) for rapid AL classification. MARLIN classification was concordant with pathology diagnoses in 18/19 (94.7%) retrospective cases profiled with nanopore sequencing, including refinement of the diagnosis in 7/19 (36.8%) cases. We further evaluated real-time MARLIN classification during nanopore sequencing in prospective patients with suspected AL, achieving an accurate methylation class prediction in less than two hours from the time of sample receipt. In summary, we show that epigenetic profiling effectively resolves the biological heterogeneity of AL and is a valid surrogate for many conventional diagnostic assays. Our machine learning- and nanopore-based framework is fast, affordable and easy to implement, making it suitable for high-tech laboratories but also in remote settings, and provides a foundation for future developments in molecular AL diagnostics.
***25 -- Mapping the topography of spatial gene expression with interpretable deep learning -- First Place Winner
Uthsav Chitra, Brian J Arnold, Hirak Sarkar, Kohei Sanno, Cong Ma, Sereno Lopez-Darwin, Shu Dan, Fenna Krienen, Benjamin J. Raphael
Spatially resolved transcriptomics technologies provide high-throughput measurements of gene expression in a tissue slice, but the sparsity of this data complicates analysis of spatial gene expression patterns. We address this issue by deriving a \emph{topographic map} of a tissue slice—analogous to a map of elevation in a landscape—using a novel quantity called the \emph{isodepth}. Contours of constant isodepth enclose domains with distinct cell type composition, while gradients of the isodepth indicate spatial directions of maximum change in expression. We develop GASTON, an unsupervised and interpretable deep learning algorithm that simultaneously learns the isodepth, spatial gradients, and piecewise linear expression functions that model both continuous gradients and discontinuous variation in gene expression. GASTON relies on a novel model of spatial gradients parametrized with a conservative, neural gradient field and is broadly applicable in spatial statistics. We show that GASTON more accurately identifies spatial domains and marker genes than existing approaches across several tissues. Moreover, GASTON identifies gradients of neuronal differentiation and firing in the brain; gradients of metabolism and immune activity in the tumor microenvironment; and gradients of calcium and developmental processes in the heart embryo. If time permits, I will also present GASTON-Mix, which improves the spatial domain identification in GASTON by integrating GASTON with a mixture-of-experts (MoE) model.
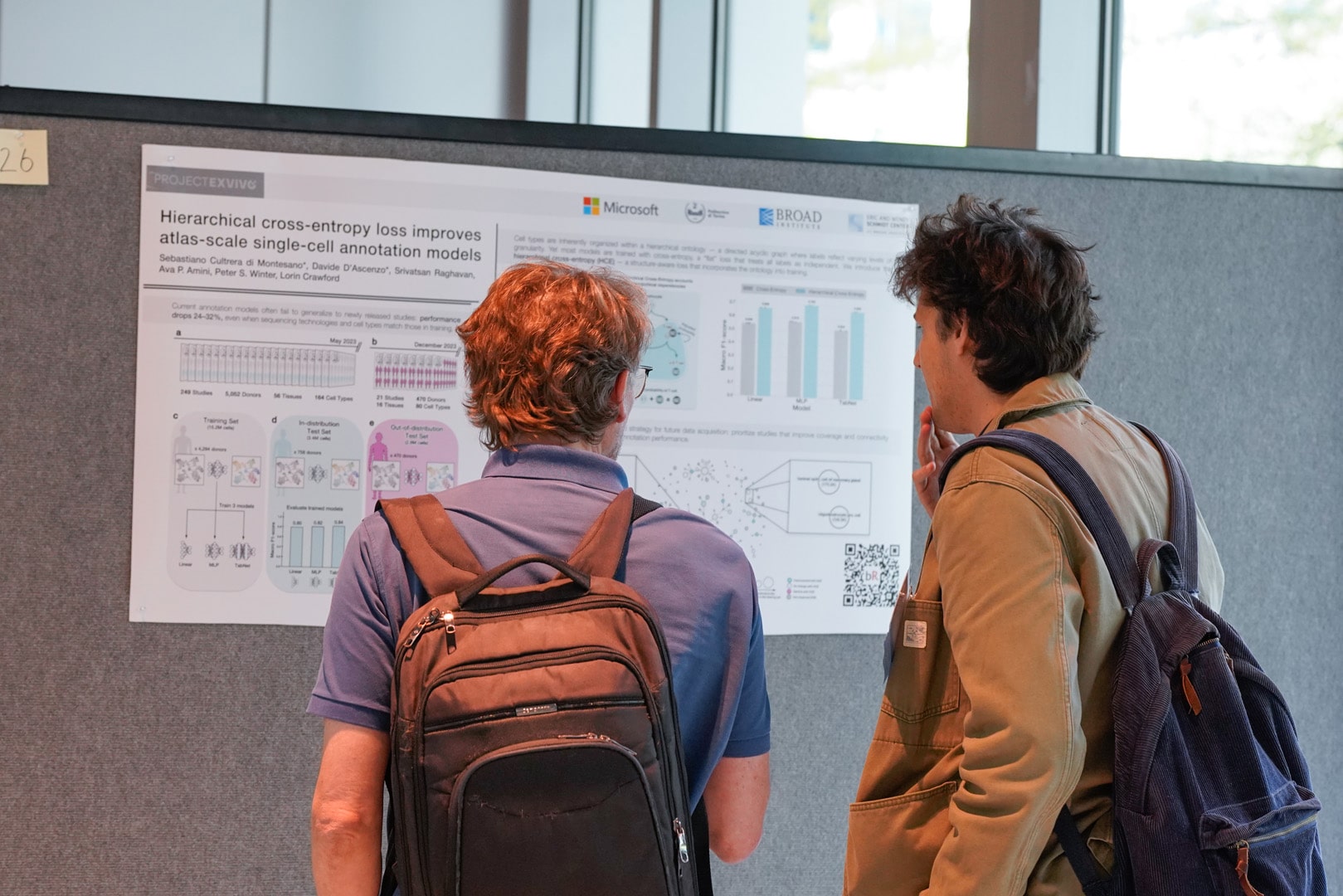
26 -- Hierarchical cross-entropy loss improves atlas-scale single-cell annotation models
Sebastiano Cultrera di Montesano, Davide D’Ascenzo, Srivatsan Raghavan, Ava P. Amini, Peter S. Winter, Lorin Crawford
Single-cell atlases now catalog millions of cells across diverse tissues, species, and experimental conditions, offering unprecedented resolution into cellular diversity and function. Accurate and robust annotation methods remain a critical first step in translating these large-scale datasets into actionable biological insights. From a machine learning perspective, cell type annotation is a multi-class classification task over a structured label space. Cell types are organized in a hierarchical ontology, where directed edges link broader categories to more specific subtypes, forming a directed acyclic graph (DAG) that encodes biological relationships among labels. Yet, most computational models—including deep learning approaches—optimize a standard cross-entropy loss that assumes flat, mutually exclusive labels, disregarding this structure entirely. We introduce a hierarchical cross-entropy (HCE) loss that aligns model training with the cell ontology. By redistributing probability mass from specific subtypes to their broader parent types, HCE encourages biologically coherent predictions—particularly in cases of label ambiguity or inconsistent granularity across datasets. To evaluate this approach, we trained three models of increasing complexity—a linear classifier, a multilayer perceptron (MLP), and a transformer-based model—on 15.2 million annotated human cells spanning 164 cell types. We then tested these models on 2.6 million newly added cells from 21 held-out studies, all sequenced with the same technology and annotated using a subset of the same labels seen in training. Incorporating HCE improved macro F1-scores by 12–15% across all models, without requiring changes to architecture or additional computational cost. These gains were consistent across model types and test datasets, and particularly pronounced for cell types embedded in densely connected regions of the ontology. Our results demonstrate the value of structure-aware training and suggest a strategy for guiding future dataset selection to further enhance model generalization.
27 -- Developmental Pattern Formation Occurs via Cue-Driven Cellular Diversification
Mayank Mangesh Ghogale, Dakota Hawkins, Alexandra Lion, Rachel Ferrigno, Sophie Bodine, Christopher F. Thomas, Nahomie Rodriguez-Sastre, Abigail E. Descoteaux, Gabriella Zambrano, James Huth, Cynthia A. Bradham
Pattern formation mechanisms remain poorly understood, particularly for complex, 3-D structures. The sea urchin larval skeleton provides an elegant, morphologically simple model to study developmental pattern formation. In this two-component system, mesodermal cells called PMCs produce the calcium carbonate skeleton in response to patterning cues that are expressed in discrete spatial regions by the adjacent ectodermal cells. Our goal is to understand the temporal and spatial mechanisms for patterning cue reception at time points 15, 18, 21, 24 and 30 hours post fertilization (hpf). To achieve this, we identified and spatially mapped the gene expression trajectories within PMC’s using single cell RNA sequencing data along with single molecule-FISH (sm-FISH) results. We employed Identify Cell states Across Treatments (ICAT), an algorithm we developed to identify PMC trajectories and marker genes therein. We then mapped the trajectory markers spatially using whole mount sm-FISH, to spatially integrate the scRNA-seq results. The sm-FISH data was analyzed using Napari, ANTS and scikit-image by first manually applying spatial landmarks to each embryo for registration, producing a spatial template, then segmenting the template into subcellular Regional Adjacency Graph (RAGs) and finally mapping the expression data onto the RAG’s. The results show that PMC trajectories map to spatially discrete locations and are temporally dynamic. At 18 hpf, each spatial region in the PMC pattern exhibits a unique combination of trajectories, suggesting that the trajectories collectively represent a spatial code for pattern formation.
28 -- Sequence-based protein models for the prediction of mutations across priority viruses
Sarah Gurev, Noor Youssef, Navami Jain, Debora Marks
Viruses pose a significant threat to human health. Advances in machine learning for predicting mutation effects have enhanced viral surveillance and enabled the proactive design of vaccines and therapeutics, but the accuracy of these methods across priority viruses remain unclear. We perform the first large-scale modeling across 40 WHO priority pandemic-threat pathogens, many of which are under-surveilled, discovering that most have sufficient sequence or structural information for effective modeling, highlighting the potential for using these approaches in pandemic preparedness. To understand the limits of current modeling capabilities for viruses, we curate 47 standardized viral deep mutational scanning assays to systematically evaluate the performance of three alignment-based models, three Protein Language Models (PLMs), and two structure-aware PLMs with different training databases. We find marked differences in performance of these models on viruses relative to non-viral proteins. For viral proteins, we find alignment-based models perform on par with PLMs though with predictable differences in which model is better for a particular function or virus depending on data available. We define confidence metrics for both alignment-based models and PLMs that indicate when additional sequence or structural data may be needed for accurate predictions and to guide model selection in the absence of available data for evaluation. We use these metrics to inform the development a confidence-weighted hybrid model that builds on the strength of each approach, adapts to the quality of data available, and outperforms either of the best alignment or PLM models alone.
29 -- Mapping functional genetic variants and disease heritability in human adipocyte villages under metabolic disease-relevant stimuli
Yi Huang, Joaquín Pérez-Schindler, Sanchari Datta, Bellis Min, Nivedita Nambrath, Mayank Murali, Hesam Dashti, Bandana Sharma, Sunita Singh Poma, Phil Kubitz, Thouis R. Jones, Melina Claussnizter
The rising prevalence of metabolic diseases represents a global public health concern, with limited understanding of pathophysiological mechanisms hindering the development of effective therapeutic approaches. Genetic variation and its interaction with environmental cues are crucial in the etiology of metabolic diseases, which are strongly linked to adipocyte dysfunction and are highly cell state- and context-specific. Here, we leverage a population-scale biobank (CellGenBank) to conduct pooled natural genetic variation screens in primary human adipocyte villages for single-nucleus transcriptomic and chromatin accessibility profiling under various disease-relevant stimuli. Processing 338k nuclei from 118 donors allowed us to identified key cell states characterized by canonical marker genes, including quiescent (PDGFRA) and proliferative (PDGFRA, CDK1, AURKB) adipose tissue-derived mesenchymal stem cells (AMSCs), structural Wnt-regulated adipose tissue-resident (SWAT; DCN, PLAC9, APOD) cells and adipogenic (ADIPOQ, PLIN1) cells, with distinct transcriptional responses to stimuli. By associating individual polygenic risk scores, we identified correlations between disease risk and shifts in cell state proportions. These states have been also mapped to metabolic disease relevant traits using single-cell heritability analysis, with body mass index highly enriched in AMSCs, whereas traits informative of metabolic health show strong enrichment in adipogenic cells and a subpopulation of SWAT cells. Furthermore, genome-wide eQTL mapping revealed hundreds to thousands of context- and cell state-specific eQTLs with potential roles in adipocyte function. Collectively, the implementation of the human adipocyte village approach coupled with single-nucleus functional genomics enabled the discovery of genetic mechanisms underlying metabolic diseases, highlighting its high therapeutic potential.
30 -- Leveraging diverse ancestry data to uncover genotype-driven transcriptomic mechanisms in adult and developing brain for psychiatric disorders
Aarti Jajoo, C. Chatzinakos, V. Balakundi, A. Aruldass, C. Wen, I. G. C. Kolmans, H. Schuler, A. Iatrou, J. E. Kleinman, K. J. Ressler, T. Bigdeli, M. J. Gandal, N. P. Daskalakis
To better understand molecular mechanisms underlying psychiatric disorders, we need improved analytic approaches for integrating large-scale genomic data with biological data representing gene transcription in the human brain. However, one critical component of individual variation - different levels of gene regulation due to genetic ancestry diversity - has not been traditionally incorporated into such analyses. To address this, we leveraged the ancestral diversity of individuals in Genotype-Expression (GEx) reference panels and GWAS to enhance the detection of transcriptome-wide association study (TWAS) signals. To investigate GEx-level choices, we trained GReX (Genetically Regulated gene expression) models using rigorously constructed subsets of a human postmortem cortex GEx panel, generated through downsampling, segregating, and mixing samples of Admixed African (AA) and European (EUR) ancestry, while considering disease status in the subset design. TWAS results were obtained by integrating these GReX models with ancestry-specific GWASs for schizophrenia (SCZ), post-traumatic stress disorder (PTSD), major depressive disorder (MDD), and bipolar disorder (BIP). Ancestry-specific predicted genes were enriched in specialized pathways involving mitochondrial functions, organelle structure, and metabolism, while shared genes demonstrated high concordance (>95%) in predictor SNP weight direction. Shared TWAS signals, obtained by integrating various GReX models with a GWAS, demonstrated high concordance while uncovering novel signals at the gene, pathway, and drug-repurposing levels. Despite lower power due to smaller cohort sizes, AA GWASs enhanced signals and alleviated noise in meta-TWAS analysis when integrated with appropriate GReX models. EUR GReX-specific TWAS pathways included corticosteroid signaling in PTSD, TGF-beta and neurotrophins in MDD, and inflammation and viral life cycle in SCZ. AA GReX-specific TWAS pathways included glutamine signaling in PTSD, proline-peptide DNA activity in MDD, and immune cytotoxicity, serotonin, dopaminergic, and phagocytosis pathways in SCZ. Finally, ancestry-specific predicted genes in the developing brain exhibited pathway enrichment like the adult brain, but showed a higher proportion of shared TWAS signals across ancestries, alongside prominent ancestry-specific signals in specialized developmental and neuronal pathways.
***31 -- CellWHISPER: Inference of contact-mediated cell-sell signaling -- Second Place
Anurendra Kumar, Nicholas Zhang, Bhavay Aggarwal, Ahmet Coskun, Saurabh Sinha
Recent advancements in single-cell transcriptomics have transformed our understanding of cellular communication by elucidating intricate interactions between cells, genes, and signaling pathways. While tools like CellChat effectively infer ligand-receptor (LR)-mediated signaling from scRNA-seq and spatial transcriptomics(ST) datasets, contact-mediated communication—particularly via gap junctions(GJ)—remains unexplored. Furthermore, existing methods leveraging spatial information for LR-mediated signaling fail to account for confounding effects from the spatial distribution of cell types, leading to high false-positive rates. Here, we present WHISPER (Workflow for HIgh-precision Spatial Proximity-mediated cEll-cell inteRactions), a statistical framework that constructs contact networks from ST data to model cell-pair connectivity and infer a tensor representing interactions across cell types and signaling gene pairs (GJ or LR). To enhance scalability, WHISPER derives an analytical null distribution, enabling efficient analysis of large datasets. Applying WHISPER to mouse brain ST data, we demonstrate superior false-positive control compared to existing methods. WHISPER identifies known contact-mediated interactions, such as astrocyte-oligodendrocyte GJ communication, and reveals novel interactions, including excitatory-astrocyte GJ coupling and excitatory-granule neuron LR signaling in hippocampus. To uncover broader communication patterns, we introduce a latent variable model that detects recurrent interaction e.g. cell types that interact using combinations of signalling gene pairs, resulting in a robust map of contact-mediated communication. Furthermore, we develop imputation strategies to extend WHISPER’s applicability to imaging-based datasets with limited gene throughput. Electrical synapses (mediated by GJs) and chemical synapses (mediated by LRs) are known to regulate each other in specific contexts. Our findings reveal a novel crosstalk between LR- and GJ-mediated signaling, particularly enriched in extracellular matrix-related pathways. Finally, we apply WHISPER to Alzheimer’s disease models, performing differential analyses to identify both local and global alterations in cell-cell communication. These results provide key insights into the interplay between electrical and chemical synapses, shedding light on their coordinated role in neural development and disease.
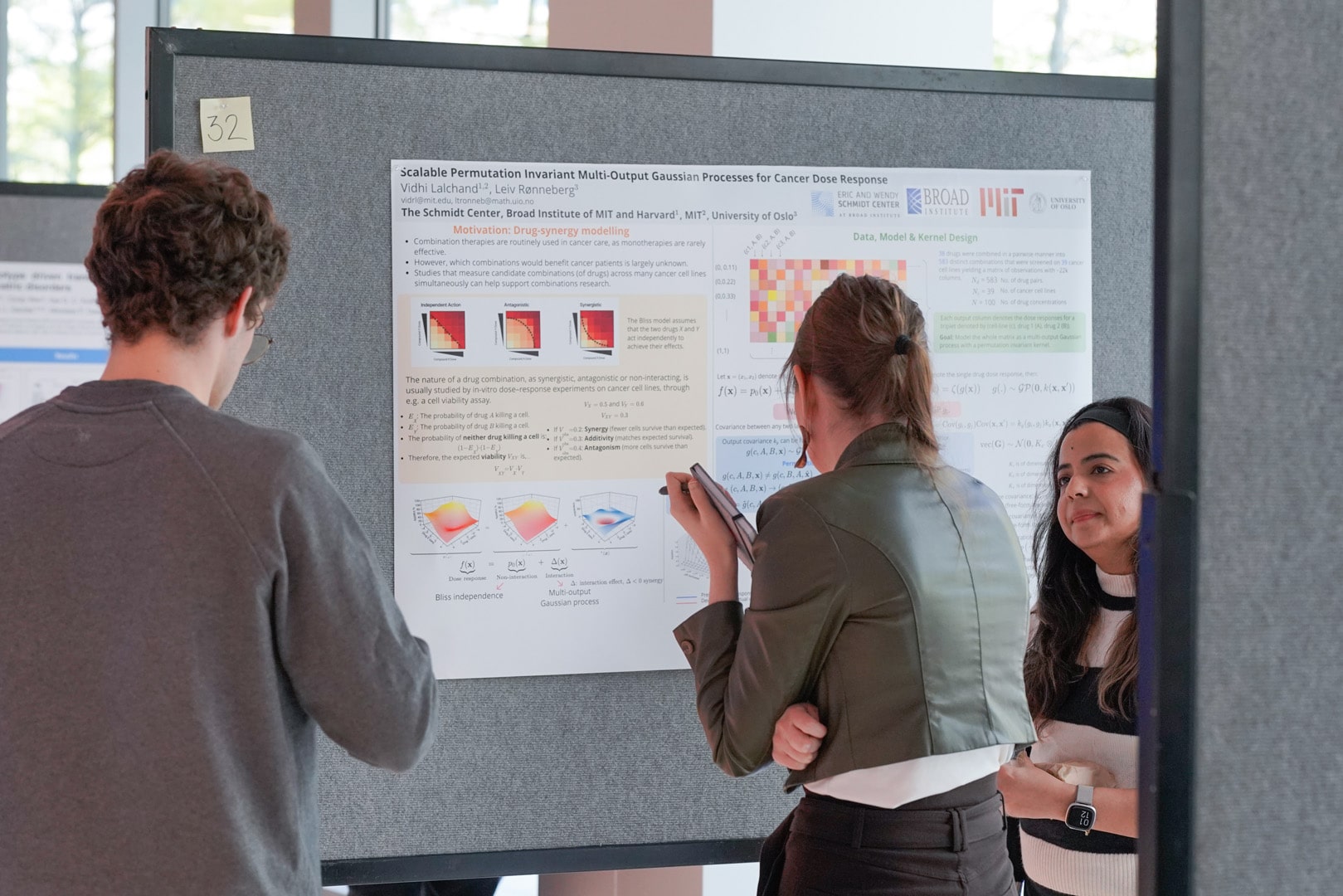
32 -- Cancer Drug Response Surface Modelling with Multi-Output Gaussian Processes
Vidhi Lalchand, Leiv Ronnenberg
Dose-response prediction in cancer is a critical step to assessing the efficacy of drug combinations on cancer cell-lines. The efficacy of a pair of drugs can be expressively modelled through a dose-response surface which outputs the viability score across a spectrum of drug concentrations for each pair of drugs in the training data. Using large in-vitro drug sensitivity screens, the goal is to develop accurate predictive models that can be used to inform treatment decisions by predicting the efficacy of given drug combination on new cancer cell lines as well as predict the effect of unseen drugs on seen cancer cell-lines. Previous work for modelling dose response surfaces precluded scalability to large datasets and did not encode cell-lines and drugs, hence, these frameworks couldn't generalise to novel combinations of drugs and cell-lines. We achieve this through the use of an upstream a deep generative model (DGM) to embed the drugs in a continuous chemical space - enabling viability predictions for unseen drugs. We use a similar approach to encode cancer cell-lines. We demonstrate the performance of our model using an open source high-throughput dataset and show that it is able to efficiently borrow information and model cross-covariance across experimental outputs where each output encodes the efficacy across a grid of concentrations of a specific drug pair on a fixed cancer cell-line.
33 -- 3D interchromosomal interactions mediate pathway-level gene regulation and shape the aging phenotype in human skin fibroblasts
Max Land, Louis Cammarata, G. V. Shivashankar, Caroline Uhler
Fibroblasts play a critical role in maintaining the extracellular matrix (ECM) integrity by sensing and responding to mechanical signals from the tissue environment. A defining characteristic of aging, the deterioration of the ECM disrupts tissue homeostasis and is implicated in conditions such as fibrosis, cancer, and other aging-related diseases. However, the factors that influence fibroblasts to change state and function during aging remain unknown. Here, we perform comparative statistical analysis of Hi-C data from healthy skin fibroblasts of a young (10 yo) and old patient (75 yo). Age-specific analysis of 3D interchromosomal contacts revealed the spatial colocalization of genes involved in ECM, TGF-β signaling, and senescent pathways in young and inflammatory pathways in old. Using transcription factor (TF) enrichment analysis, we identified potential TF regulatory subnetworks (transcription factories) within spatially colocalized genes. Computational analysis of pathway-specific stimulation experiments (e.g., TGF-β, cytokines) between old and young fibroblasts confirmed functional differences in phenotype aligning with the presence of transcription factories. Collectively, our results suggest a pathway-level mechanism of gene regulation in which the 3D chromatin organization of fibroblasts facilitates spatial clustering of pathway genes, poising the cell toward specific phenotypes. This finding provides insights into how the interplay between ECM changes, mechanical signaling, and 3D chromatin directly shapes the ageing phenotype. In addition, this gene regulatory mechanism could potentially be targeted to develop novel therapeutic strategies to treat ageing and other fibroblast-related diseases.
34 -- Integrating Mechanistic RNA Dynamics with Machine Learning for Gene Regulatory Network Inference in Multimodal Single-Cell Data
Alexander Aivazidis, Tara Chari, Matthew Levine, Luca Pinello, Cameron Smith
Multimodal single-cell experiments enable the simultaneous measurement of chromatin state, and nascent and mature mRNA across thousands to millions of cells, providing an unprecedented view of transcriptional processes over development and perturbation. These data offer a unique opportunity for mechanistic inference of how DNA state changes propagate through RNA processing and how regulatory interactions govern transcriptional dynamics. However, classical models of stochastic RNA dynamics either assume independent gene regulation or scale poorly beyond small, predefined networks. Machine learning approaches, while flexible, often rely on black-box predictions and ad hoc data transformations, limiting mechanistic interpretability. Here, we introduce a framework that integrates continuous mechanistic models of stochastic RNA dynamics with machine learning to infer gene regulatory networks. This approach enables scalable Bayesian inference of gene regulatory dynamics governing DNA and RNA processing across neural and immune cell differentiation as well as under unseen genetic interventions. Beyond predicting cell fate, our method reveals regulation through transcription factor interactions and binding, and the downstream effects on RNA processing kinetics.
35 -- Unraveling Tissue Assembly with Hierarchical Transformers
Dongshunyi Li, Sandeep Kambhampati, Qiyu Gong, David Benjamin, Ebtisam Alshehri, Yerdos Ordabayev, Mehrtash Babadi, Fei Chen, Caroline Uhler
One of the fundamental questions in biology is how cells interact and assemble into tissues, organs and ultimately the entire human body. While numerous studies have advanced our understanding of this process, only recently has it become possible to investigate the rules of tissue organization in a data-driven manner. High-throughput spatial transcriptomics technologies have transformed molecular biology by enabling the measurement of molecular features at scale directly within tissue context. The resulting large-scale, high-dimensional spatial data provide unique opportunities to develop models that learn the principles governing tissue assembly. We introduce TISU (Tissue-Interaction & Spatial Understanding), a transformer-based model designed to identify the multi-hierarchical spatial organization of cells and the interaction mechanisms among them. Inspired by the concept of compartmentalization in multicellular organisms, TISU captures spatial structures from the cellular level to tissue module level, learning interaction rules within and across these organizational levels. By training on millions of cells, our model develops a systematic and comprehensive understanding of tissue assembly and holds the potential to reconstruct tissues from individual cells in silico. Through rigorous evaluation, we demonstrate that TISU exhibits generalization capabilities to unseen tissue sections, accurately reconstructing missing cells and spatial modules. Additionally, the attention maps generated by the model reveal biologically meaningful spatial interactions and provide insights into tissue organization. These results highlight the promise of TISU as a powerful tool for advancing our understanding of tissue assembly and organization.
36 -- No poster
37 -- CelloType: a unified model for segmentation and classification of tissue images
Minxing Pang, Kai Tan
Cell segmentation and classification are critical tasks in spatial omics data analysis. Here we introduce CelloType, an end-to-end model designed for cell segmentation and classification for image-based spatial omics data. Unlike the traditional two-stage approach of segmentation followed by classification, CelloType adopts a multitask learning strategy that integrates these tasks, simultaneously enhancing the performance of both. CelloType leverages transformer-based deep learning techniques for improved accuracy in object detection, segmentation and classification. It outperforms existing segmentation methods on a variety of multiplexed fluorescence and spatial transcriptomic images. In terms of cell type classification, CelloType surpasses a model composed of state-of-the-art methods for individual tasks and a high-performance instance segmentation model. Using multiplexed tissue images, we further demonstrate the utility of CelloType for multiscale segmentation and classification of both cellular and noncellular elements in a tissue. The enhanced accuracy and multitask learning ability of CelloType facilitate automated annotation of rapidly growing spatial omics data.
38 -- Multi-marginal Schrödinger Bridges with Iterative Reference Refinement
Yunyi Shen, Renato Berlinghieri, Tamara Broderick
A key challenge in automated biomedical discovery is ensuring that AI systems make interpretable decisions grounded in comprehensive literature review. To this end, scientists often undertake a series of actions, reading and reasoning over documents from multiple sources and deriving new insights. We build an agent system that conducts long chain-of-thoughts reasoning with retrieval for successfully answering such complex questions. Specifically, we formalize key characteristics in the complex reasoning traces of multi-document reasoning on biomedical literature, develop an interface for biomedical researchers to execute complex reasoning in collaboration for data collection, and train and evaluate the agent system to perform the retrieval and reasoning.
39 -- No poster
40 -- Towards detecting molecular quantitative trait loci on neurotypical population
Maria Skoularidou, Nikolaos Daskalakis
There is a vast literature that has been evolved over the past few decades focusing onthe detection of molecular quantitative trait loci (QTLs). These quantitative traits arecharacteristics that depend on inherited factors and whose intensity is affected by interactions between genes and environmental factors. In the present work we shall focus on the identification of such eQTLs across chromosome, using linear methods, based on postmortem data of neurotypical individuals of European origin.
41 -- No poster
42 -- Diffusion Model for Compositional Representation of Subcellular Structures in Eukaryotic Cell Division
Sophie Sun, Athanasios Litsios, Lance Chao, Rahul G. Krishnan
In human cells, abnormalities in the cell cycle are hallmarks of diseases such as cancer. However, understanding how subcellular structures and their respective functions dynamically change in space and time during the cell cycle remains challenging. Live-cell imaging techniques, such as fluorescence microscopy, can analyze the spatiotemporal dynamics of subcellular structures as a function of the cell cycle. However, limitations related to spectral overlap do not allow the simultaneous visualization of more than a few structures within a single cell. This prevents us from transitioning towards integrated models of intracellular organization, whereby complex physical and functional interactions between multiple structures are resolved in space and time. Here, we present YeastDiff, a computational approach for addressing limitations of simultaneously monitoring multiple structures within a single cell. YeastDiff is a cell-visualization tool that leverages conditional diffusion models (CDMs) and compositionality to generate high-resolution images of multi-subcellular structures. Trained on millions of live-cell images each depicting one of ~4000 different yeast proteins at a specific cell cycle stage, YeastDiff learns joint embedding of both the cell stage and gene identities to guide single and two-protein image generation. In addition to being able to generate biologically realistic single-cell images depicting each possible pairwise combination of subcellular structures, we are currently improving the compositional capabilities of YeastDiff to accurately represent spatial interactions between multiple proteins. Collectively, YeastDiff addresses limitations in conventional fluorescence microscopy, enabling the study of the cell cycle-related dynamics of subcellular structures in an integrated fashion. This computational framework opening new avenues for investigating cell cycle regulation and disease mechanisms.
43 -- Variation and regulatory mechanisms of the small RNA transcriptome across human tissues
Petar Stojanov, Tim Coorens, Juan Carlos Fernandez del Castillo, Scott Steelman, Sarah Young, Chad Nussbaum, Gad Getz, Kristin Ardlie, François Aguet
Standard RNA-sequencing protocols exclude small RNAs and thus preclude the study of thousands of small noncoding RNAs with essential roles in the post-transcriptional regulation of gene expression. Here, present the characterization of small RNAs across 16,814 samples, 47 tissue sites and 978 donors in the GTEx Project. We quantified the expression of a total of 41,458 small RNAs, including microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), transfer RNAs, small nuclear RNAs, small nucleolar RNAs, Y RNAs, and others. We used supervised classification to identify putative novel RNAs not present in references, and detected 57 novel high-confidence miRNAs. We mapped QTLs in cis and trans, identifying 100s to 1000s of cis-eQTLs for each small RNA species. Among them, we discovered two trans-QTLs for tRNAs, corresponding to splice QTLs in TRMT1 and DTWD1, which alter the base editing activity of their respective target tRNAs. To investigate the propagation of genetic effects on coding genes through miRNAs, we fine-mapped and co-localized SNPs that affect miRNA expression in cis and mRNA expression in trans. Through mediation analysis we confirm miRNA-mRNA pairs are causally related, which we further corroborate through seed-pairing and conservation analysis. Notably, the tissue specificity of miRNA expression is reflected in the tissue specificity of complex traits co-localizing with miRNA eQTLs. For example, we identify an interaction between miR-5683 and ODAD1 in the cerebellum, a gene involved in the motility of glial cells. Furthermore, we found that this shared causal variant colocalized with the GWAS trait of tau levels. In summary, we demonstrate the importance of characterizing the full spectrum of small RNAs, which play critical roles in the regulation of gene expression, including in development and disease.






















.jpg)